-
Role of Ribosome Recycling Factor as a Ribosome Releasing Factor
Yoshio Inokuchi, Fabio Quaglia, Akikazu Hirashima, Yoshihiro Yamamoto, Hideko Kaji, and A. Kaji
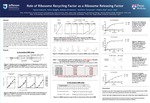
The objective of this presentation is to study the in vivo actions of ribosome recycling factor (RRF) and compare them with those found in vitro. RRF is known to catalyze three separate reactions: release of tRNA and mRNA from the post-termination complex (PoTC), and splitting of the ribosome of the PoTC. To study the mechanism of RRF reaction in vivo, we used E. coli harboring temperature sensitive (ts) RRF and assayed by following downstream reading of translationally coupled ORF. At the non-permissive temperature, ribosomes remain on the termination codon of the junction sequence of coupled ORFs and translate downstream ORF lacking a Shine-Dalgarno (SD) sequence. The readings were in all three frames due to thermal frameshift at the termination codon. When upstream ORF was short, translation of the downstream reading was abolished, suggesting that the ribosomes released by RRF are moving toward the SD sequence of the upstream ORF. The thermal frame shift at the stop codon was also stopped by the nearby upstream SD sequence. Our data suggest that the ribosome-bound mRNA may take a secondary structure around the junction sequence. This structure can affect the reading of downstream ORF. For in vitro studies, we used mRNA that incorporated different radioactively labeled amino acids based on the frameshift at the junction sequence, UAAUG, of two translationally coupled ORFs. In the absence of RRF, the ribosome stayed on the mRNA and translated in frame with the termination codon UAA. In the presence of RRF, amino acid incorporation occurred in frame with the start codon AUG. This suggests that RRF releases the ribosome from UAA and the released ribosome binds to AUG and begins translation. With the use of tethered, unsplittable ribosomes (Ribo-T) in the in vitro system, we showed that complete ribosomal splitting is not required for the action of RRF. Therefore, the main role of RRF in the ribosome recycling reaction appears to be the release of ribosomes from mRNA.
-

Understanding and Impact of Negative Direct-to-Consumer BRCA Test Results: A Pilot Study
Kristen Pauley, Zohra Ali-KhanCatts, Rachael Brandt, Cristina Nixon, and Robert Sterling
Many people pursue direct-to-consumer (DTC) DNA testing to learn about health risks. While DTC testing for breast cancer genes BRCA1 and BRCA2 may provide vital information to those testing positive, it is unclear whether individuals testing negative understand the implications of these results. Furthermore, it is unknown how such findings may influence future cancer risk management. Ninety-six individuals who underwent BRCA testing through the DTC service, 23andMe, completed a survey designed to assess their understanding of a negative result and how this result may impact cancer risk management. Respondents were recruited via social media sources Reddit and Facebook. All participants tested negative for the three BRCA variants analyzed by 23andMe: BRCA1 185delAG, BRCA1 5382insC, and BRCA2 6174delT. Six questions assessed understanding of the limitations of the testing. Results indicated that 77.1% of respondents answered five or more questions correctly, confirming participants adequately understood testing limitations. While 92% of participants reported their results would not alter the frequency of their cancer screenings, analysis of variance indicated that lower understanding of testing limitations was associated with less clarity about how to proceed with cancer risk management, F (2, 93) = 5.98, p < .05. Analyses were conducted to identify factors contributing to overall understanding scores. Significant correlates of understanding were age and education level. Participants’ perceived risk played a role in both their understanding and future cancer screening behavior. This observation necessitates further study.
-

Urokinase Plasminogen Activator Expression is Regulated by p53 Harboring the Lung Cancer-Specific Mutation V157F
Julie Barta, MD; Kristen Pauley; and Steven B. McMahon, PhD
OBJECTIVES
- To define the mutant p53-regulated transcriptome of lung cancer cells with alterations at V157 and R158 in the p53 tumor suppressor.
- To determine the biological effects of lung-enriched p53 mutations in lung cancer cells.
Printing is not supported at the primary Gallery Thumbnail page. Please first navigate to a specific Image before printing.



